
News
-

COVID Catalyst Fund supports work on rapid serology test at UC Santa Cruz
Biomolecular engineer Nader Pourmand is evaluating the use of a novel nanosensor to provide rapid, low-cost, and accurate serology tests for coronavirus antibodies May 14, 2020 | Tim Stephens | UCSC Researchers at UC Santa Cruz are testing a novel nanosensor technology that could provide rapid, low-cost serology tests with high sensitivity for detecting and…
-

Dr. Salama receives NIH support to research normal and disordered brain development
IBSC | Catharina Lindley | May 7, 2020 Dr. Sofie Salama received an R01 research grant from the National Institute of Mental Health to study molecular and cellular defects that underlie complex neurodevelopmental diseases like autism and schizophrenia. Understanding these mechanisms will help in the development of approaches to prevent or ameliorate these conditions. Multiple…
-

New DNA technology leads to DNA match in Daralyn Johnson murder case
Idaho News 6 | Jessica Taylor | May 6, 2020 Cutting edge DNA technology led to a match and arrest in the Daralyn Johnson case. Boise State professor and director of the Idaho Innocence Project Greg Hampikian had a critical hand to play in the discovery. Hampikian knows DNA; he’s been studying samples of it…
-
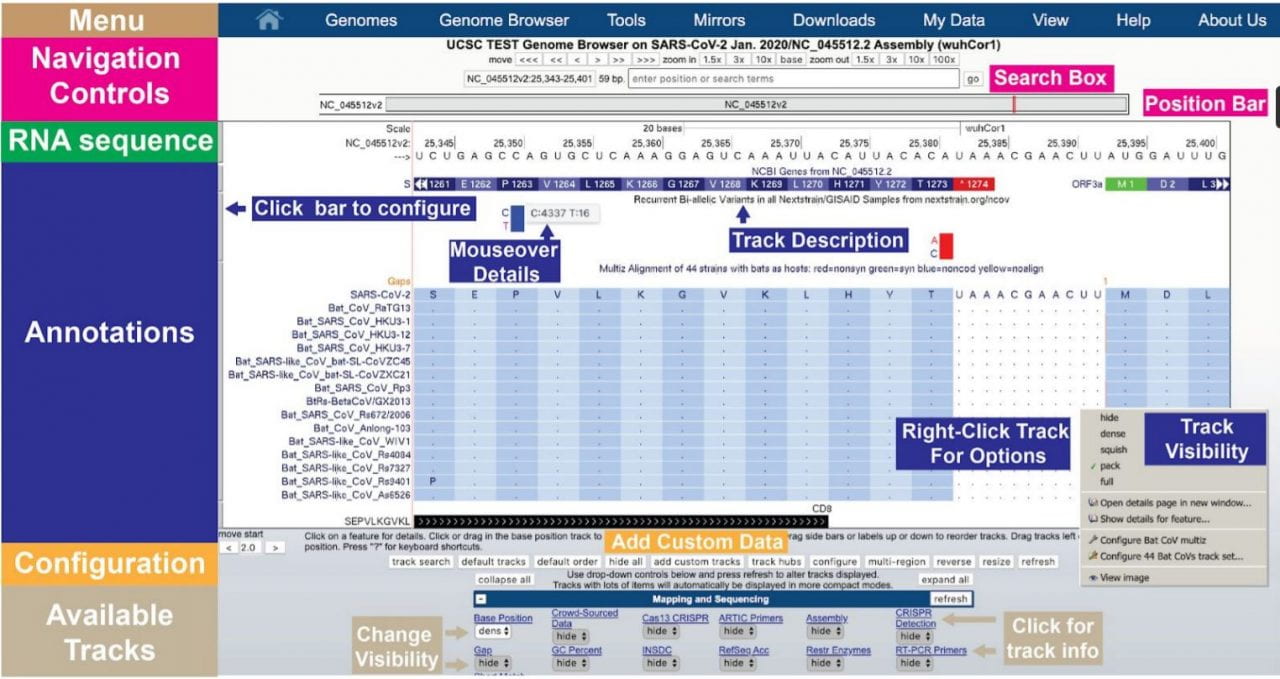
The UCSC SARS-CoV-2 Genome Browser
The UCSC Genome Browser features a Coronavirus Browser to help researchers visualize the coronavirus genome that causes COVID-19. Refer to the tutorial and video here: http://bit.ly/3qzPyQd. UCSC’s coronavirus browser maps gene and protein alignments, global variants, and even the genomes of variants from other species like bats and pangolins. It draws on sequencing data from around the world,…
-
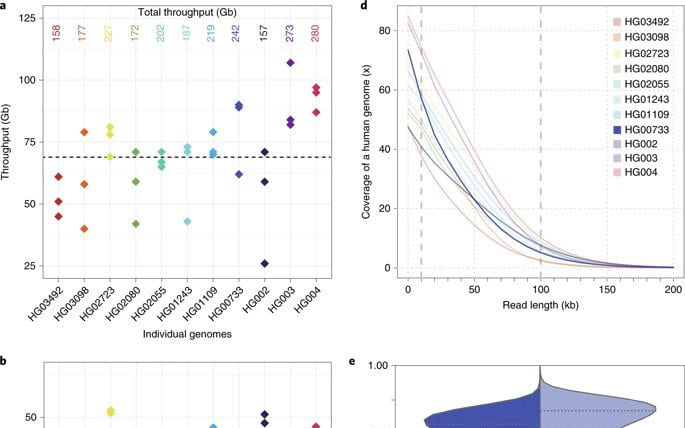
Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes
Abstract De novo assembly of a human genome using nanopore long-read sequences has been reported, but it used more than 150,000 CPU hours and weeks of wall-clock time. To enable rapid human genome assembly, we present Shasta, a de novo long-read assembler, and polishing algorithms named MarginPolish and HELEN. Using a single PromethION nanopore sequencer and…
-

UC’s CITRIS launches COVID-19 response with 25 awards for innovative technologies
Initial round includes potential “game-changing” innovation for diagnostics, therapeutics, and mitigation
-

Genome of beloved sea otter Gidget now available for browsing
UC Santa Cruz Genomics Institute | May 13, 2020 A sea otter genome browser — featuring the Monterey Bay Aquarium’s beloved Gidget – is now available to the public. The visualizable genome for the Southern sea otter, Enhydra lutris nereis, comes following work by the National Center for Biotechnology Information (NCBI) and UC Santa Cruz software bioinformaticians to make available the…
-

Team reveals genomic history of ancient civilizations in the Andes
The study included samples from the 15th-century Incan site of Machu Picchu. (Photo by Andrea Schell) UCSC | May 07, 2020 | Stephanie Dutchen An international research team has conducted the first in-depth, wide-scale study of the genomic history of ancient civilizations in the central Andes mountains and coast before European contact. The findings, published online…
-
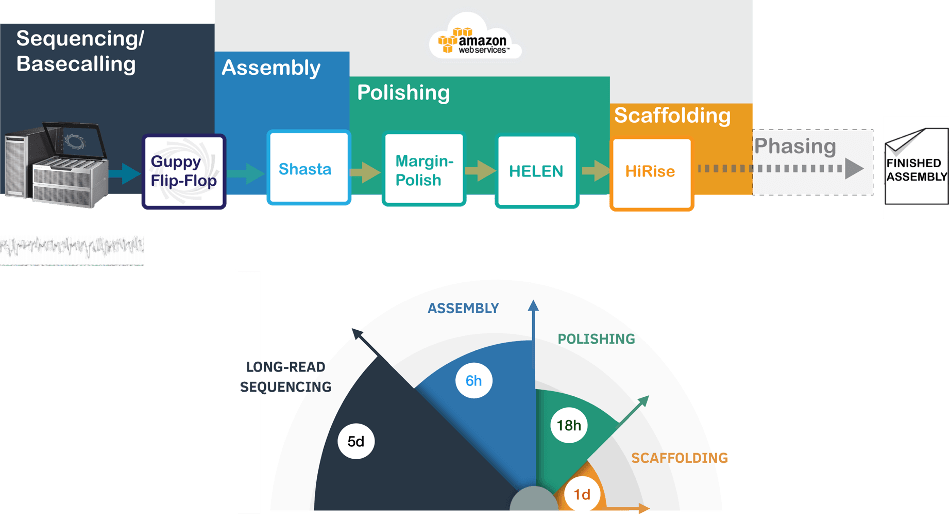
Making nanopore sequencing and de novo assembly fast and efficient
De novo assembly of human genomes using long reads has significant resource overhead. In our recent work, we demonstrate nanopore sequencing and a novel de novo assembly tool Shasta-MarginPolish-HELEN to achieve the de novo assembly of eleven human genomes in nine-days Kishwar Shafin | University of California, Santa Cruz | May 4, 2020 When choosing…
-

May 4th data release for SARS-CoV-2 genome browser
UCSC Genome Browser | May 4, 2020 We are pleased to announce our second data release for the coronavirus genome browser. Much like our first release, this includes a variety of annotation types such as CRISPR tracks, protein structure and interaction tracks, immunology tracks, and a 119-way multiple sequence alignment. One new track we would…
-
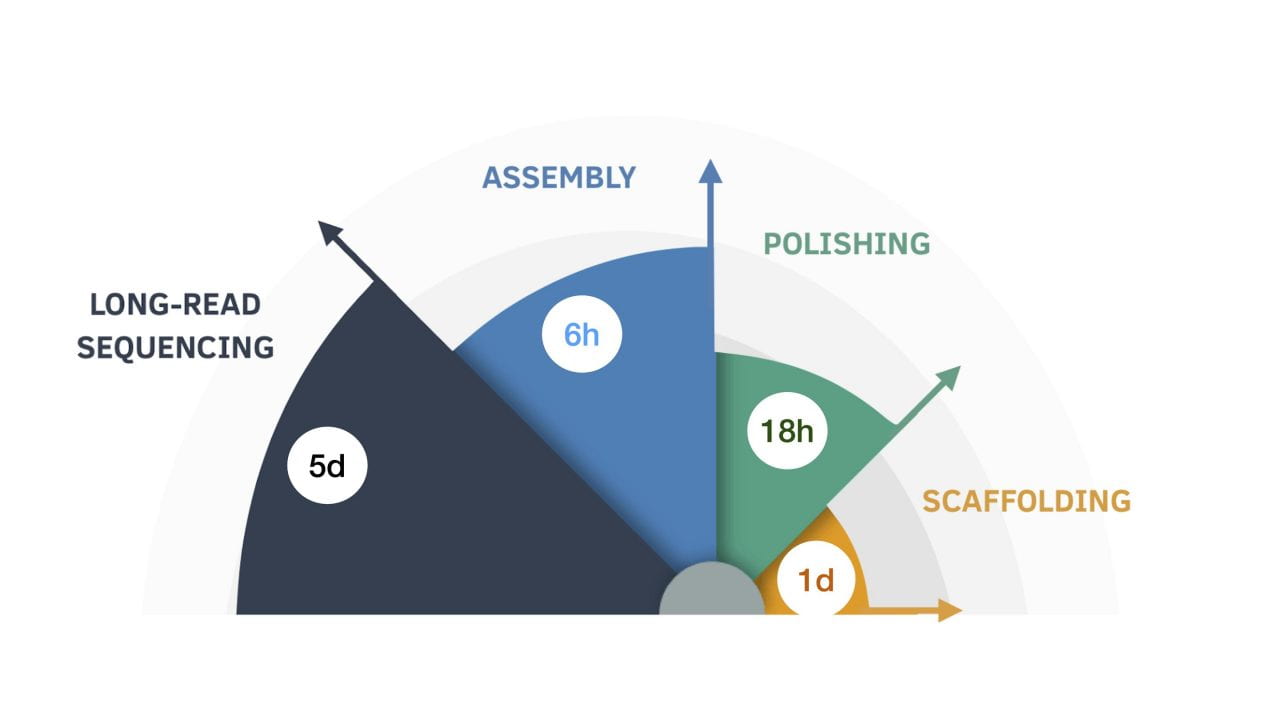
Eleven human genomes sequenced and assembled in nine days
In this diagram, the nine-day genome assembly process is broken down by length of time for each step. University of California – Santa Cruz | May 4, 2020 It’s only been three years since UC Santa Cruz researchers proved that long-read human genome assembly using the same nanopore technology developed on campus could be done at all.…
-
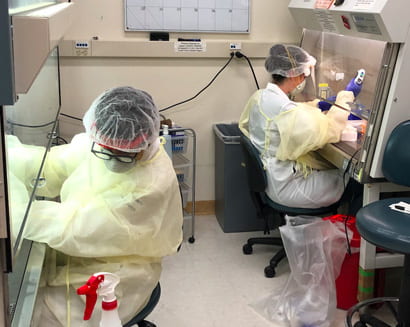
UC Santa Cruz diagnostic lab to begin coronavirus testing May 1
The campus lab will be conducting tests for coronavirus infections for the Student Health Center and local medical providers