Publications
View our full publication list on Google Scholar
Featured Publications
-
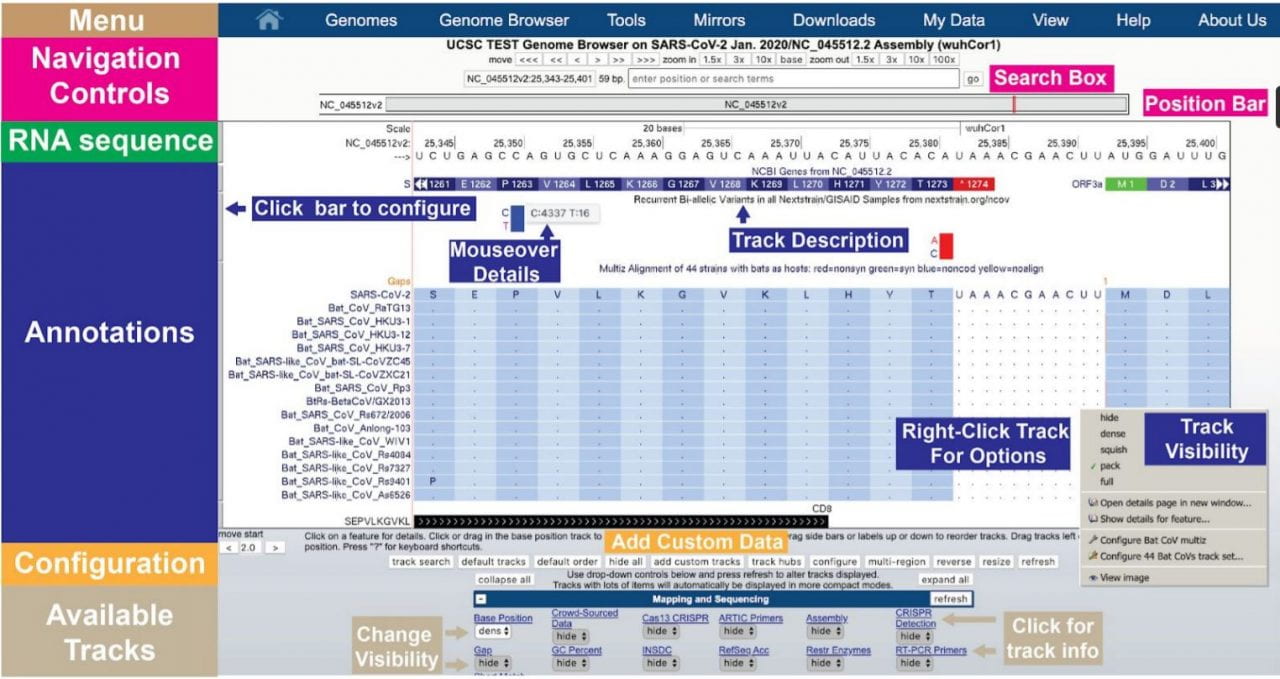
The UCSC SARS-CoV-2 Genome Browser
The UCSC Genome Browser features a Coronavirus Browser to help researchers visualize the coronavirus genome that causes COVID-19. Refer to the tutorial and video here: http://bit.ly/3qzPyQd. UCSC’s coronavirus browser maps gene and protein alignments, global variants, and even the genomes of variants from other species like bats and pangolins. It draws on sequencing data from around the world,…
-
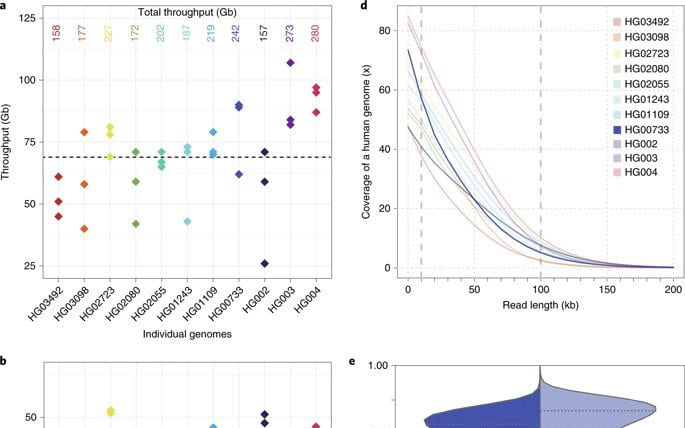
Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes
Abstract De novo assembly of a human genome using nanopore long-read sequences has been reported, but it used more than 150,000 CPU hours and weeks of wall-clock time. To enable rapid human genome assembly, we present Shasta, a de novo long-read assembler, and polishing algorithms named MarginPolish and HELEN. Using a single PromethION nanopore sequencer and…
-
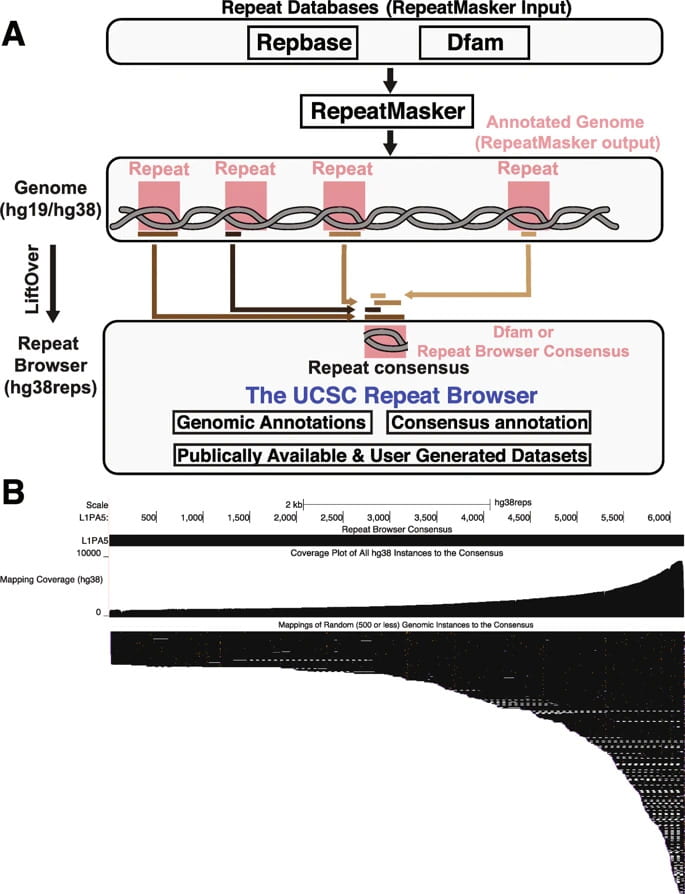
The UCSC Repeat Browser allows discovery and visualization of evolutionary conflict across repeat families
Mobile DNA | 31 March, 2020 Jason D. Fernandes, Armando Zamudio-Hurtado, W. James Kent, David Haussler, Sofie R. Salama, Maximilian Haeussler Abstract Background Nearly half the human genome consists of repeat elements, most of which are retrotransposons, and many of these sequences play important biological roles. However repeat elements pose several unique challenges to current bioinformatic analyses and visualization tools, as short repeat sequences can map to…